Instruction for HPC setup in Göttingen
In these instructions we explain how to setup a working group typical computing environment on the Göttingen HPC cluster.
There are essentially two options to work with NGSolve (and related packages) on the cluster:
- Use existing environment modules that provide the software you need. We provide some basic installations of
NGSolvefor example. If you need more specialised installations... - Install software from sources yourself (and provide them as user modules).
To get started please first read the section on the cluster login. Afterwards decide on one of the two previous options (probably option 1 is easiest to start with). If you want to use existing modules, read the section on working grouping modules. Otherwise continue with the installation instructions.
Before you ask: There will be no X-forwarding and hence no standard Netgen GUI when you run jobs through the queue. However, you can store your solutions (via pickle or VTK) to visualize on an interactive node or a different machine, e.g. your local machine.
HPC cluster login
Create a GWDG HPC account, cf. [5]. In the following we assume that your username is cinderella. The basic steps for the setup are as follows:
- For users with an existing GWDG account a short email to hpc-at-gwdg.de suffices to activate this account for the use of the HPC facilities.
- Upload your SSH public key to the gwdg website as described here [5]. (This reference also describes how to create an SSH-key in case you don't have one yet).
- Access to the frontend nodes is only possible from the GÖNET. To connect from outside to the GÖNET one could set up a VPN connection to the GÖNET. Alternatively, one can connect to the login node via
ssh login-mdc.hpc.gwdg.de -l cinderella -i ~/.ssh/id_rsa, where.ssh/id_rsais the location of your private key.
From there on login to the interactive node gwdu103 (it supports avx and avx2 vectorization), e.g. with (on unix-like systems)
ssh cinderalla@gwdu103.gwdg.de
Once logged in, we define -- for this session -- a parent directory for the following installations, e.g. your home directory:
export BASEDIR="/usr/users/cinderella"
Using pip/conda modules only
The easiest way to run NGSolve on the HPC cluster is probably using a pip/conda installation. This may not work for more advanced/derived settings with ngsxfem/ngstrefftz/ngsMumps/..., but for the basic stuff with NGSolve it should do the trick.
First, setup a configuration. To this end, call:
module load miniforge3
conda create -p /usr/users/cinderella/conda-envs/ngsolve-pip python=3.12 conda
This will create a new conda environment in /usr/users/cinderella/conda-envs/ngsolve-pip (exchange cinderella with your user name) using python3.12.
Now, you can log into this environment whenever you want with
module load miniforge3
source activate /usr/users/cinderella/conda-envs/ngsolve-pip
Within that environment you can now install whatever you need (and is available through pip), e.g.
pip install ngsolve numpy packaging matplotlib line-profiler
A simple slurm batch file to use that environment could then look like this (further explanations on the options below or on the GWDG HPC pages):
#!/bin/bash
#SBATCH -p medium
#SBATCH -t 0:24:00
#SBATCH -n 2
#SBATCH --mem 16GB
#SBATCH -N 1
module load miniforge3
source activate /usr/users/cinderella/conda-envs/ngsolve-pip
cd tough_benchmark
python benchmark_problem_83.py
Using existing modules
To load existing software you can use the existing environment modules. To see what is available run (after logging in)
module avail
To add more packages from the user level, call
module load USER-MODULES
This will add especially modules from our working group (in /cm/shared/cpde).
Now, you can check again for available packages. Assume you are looking for NGSolve. Try
module avail ngsolve
This will display different available versions. When trying to load ngsolve right away with e.g. module load ngsolve/mpi you will get an error that some dependencies are not loaded yet. If you want to check for all dependencies, call
module show ngsolve/mpi
and load all packages (in the corresponding versions) that are appear in the output. Scan for prereq_any("... in the output. Afterwards load the corresponding packages, e.g.
module load python/3.9.0
If you have done this for all dependencies you can load ngsolve
module load ngsolve/mpi
Now, you are able to load the desired package and can continue reading how to set up a run on the HPC batch system slurm.
Installation on the GWDG HPC cluster
In this section we explain installation and setup of numerical software, especially NGSolve on the GWDG HPC cluster, see also [1,2,3,4] for some general information from the GWDG on the use of the cluster.
See also below.
Before you ask 2: These instructions still miss a few points:
- Compared to the instructions below using an installation from source, somewhat simpler setups using
singularityordockerimages would be possible. We may add corresponding instruction in the future. Note that the provided environment modules already offer a convenient way to use some standard packages without the need for a manual installation. - The following instruction do not explain how to setup
ngsxfemon the cluster. As this is extremely similar to theNGSolvesetup, we leave it open for now. The same holds for the (even simpler) case ofregpy.
Why building from source?
We note that there is the possibility to use conda to install NGSolve also on the cluster. Similarly, singularity and docker (with singularity) containers could be used. However, both ways to install NGSolve cannot make perfect use of the underlying architecture, e.g. avx or avx2 simd-vectorization would not be used. Instead we explain how to build NGSolve from sources here. But careful: when using the underlying architecture specification (DUSE_NATIVE_ARCH=ON below), we need to make sure that the visible cpu instruction set at build time is compatible with the machines that you later on run NGSolve on.
In case that you are scared of the lengthy instructions below, you can use the shortcut of using a script. The first steps however (logging in, setting up directory structure, fetching the sources and configuring the modules directory) should be done (once) before.
Basic structure for installations
For the following installations we recommend to use the following structure:
$BASEDIR
├── src
├── build
├── packages
└── modules
Create those (empty) directories if they don't exist yet:
cd $BASEDIR
mkdir -p src build packages modules
The idea of these directories is:
- in
srcyou put the sources of individual packages - in
buildyou put the build directories for individual packages in possibly different versions - in
packagesyou install those packages - in
modulesyou define environment settings to "activate" the corresponding installations
We created the directory modules directory. To prepare for adding new modules here and to make sure that module avail finds our new module, we have to add this directory to the MODULEPATH so that lmod can find it. To do this for one session, just do
export MODULEPATH="$MODULEPATH:${BASEDIR}/modules"
To have this set up on every login, add this command to your shells init script (for bash this is ~/.bashrc), e.g. by
echo "export MODULEPATH=\"\$MODULEPATH:${BASEDIR}/modules\"" >> ~/.bashrc
Next, we explain how to install NGSolve in this setting.
NGSolve
Fetching sources or specifying commit
We start by cloning the sources of NGSolve [1]If you want to reproduce some results from older version this is where you should adjust to the corresponding version of NGSolve.:
cd $BASEDIR
git clone https://github.com/NGSolve/ngsolve src/ngsolve
cd src/ngsolve
git submodule update --init --recursive
cd $BASEDIR
At this point, you can take the shortcut and build and install from the provided script. However, we still recommend to follow the instructions in the next sections so that you know what is happening.
Build step
For the underlying ngsolve version and build options[2]for example serial or MPI parallel. we fix a name, e.g.:
export BUILDTYPE="serial-`cd $BASEDIR/src/ngsolve && git describe --tags`"
Next, we have to decide for a python version that we want to use. To be flexible in the remainder we define corresponding environment variables:
export PYTHONMODULE="python/3.9.0"
export PYTHONVER="3.9"
Alternatively, you may also use intel/python/2018.0.1 and 3.6 rather than python/3.9.x and 3.9because this is optimized for intel machines. However, a.t.m. it seems that when using intel/python the module needs to loaded also before submission of the job (which shouldn't be the case usually).
We load the modules that we want to make use of[3]Note that (most of) these will be required to be loaded when running ngsolve later. :
module load ${PYTHONMODULE}
module load gcc/9.3.0
module load cmake
module load intel-parallel-studio/cluster.2020.4
We will put all built packages in the packages and build from a corresponding directory in build. We clean directories with same name and switch to the build directory:
mkdir -p $BASEDIR/packages/ngsolve
cd $BASEDIR/packages/ngsolve
rm -rf ${BUILDTYPE}
mkdir ${BUILDTYPE}
mkdir -p $BASEDIR/build/ngsolve
cd $BASEDIR/build/ngsolve
rm -rf ${BUILDTYPE}
mkdir ${BUILDTYPE}
cd ${BUILDTYPE}
We are now in the new (or cleaned) directory to build ngsolve. To this end, we call[4]you may want to adapt the build options
cmake \
-DUSE_GUI=OFF \
-DUSE_UMFPACK=ON \
-DUSE_MUMPS=OFF \
-DUSE_HYPRE=OFF \
-DUSE_MPI=OFF \
-DCMAKE_BUILD_TYPE=RELEASE \
-DCMAKE_INSTALL_PREFIX=${BASEDIR}/packages/ngsolve/${BUILDTYPE} \
-DCMAKE_CXX_COMPILER=g++ \
-DCMAKE_C_COMPILER=gcc \
-DUSE_MKL=ON -DMKL_ROOT=${MKLROOT} \
-DUSE_NATIVE_ARCH=ON \
-DCMAKE_CXX_FLAGS="-ffast-math" \
${BASEDIR}/src/ngsolve/
If cmake preparations run through, we can build the package with a few cores, e.g.:
make -j12
make install
cd ${BASEDIR}
To run ngsolve with this installation you have to adjust environment variables such as PYTHONPATH, PATH, INCLUDE_PATH, LD_LIBRARY_PATH or NETGEN_DIR correspondingly and make sure that the required modules are preloaded. This can be automated. For this we use lmod, the same tool that we already used before to load gcc, mkl, cmake and python.
Add your installation as a module
Each file in the modules directory corresponds to an environment module that can loaded. Within the directory different versions may be stored.
A package file ${BUILDTYPE} refering to the corresponding NGSolve installation in ${BASEDIR}/packages/ngsolve/${BUILDTYPE} may look as follows:
#%Module1.0################################################# -*- tcl -*-
#
# GWDG Cluster gwdu10X
proc ModulesHelp { } {
puts stderr tSets up environment for NGSolve
}
module-whatis NGSolve
prereq gcc/9.3.0
prereq ${PYTHONMODULE}
prereq intel-parallel-studio/cluster.2020.4
prereq intel/mkl/64/2020/2.254
conflict ngsolve
set base ${BASEDIR}/packages/ngsolve/${BUILDTYPE}
setenv NETGEN_DIR $base/bin
setenv NGSCXX_DIR $base/bin
prepend-path PATH $base/bin
prepend-path INCLUDE_PATH $base/include
prepend-path LD_LIBRARY_PATH $base/lib
prepend-path LD_PRELOAD ${MKLROOT}/lib/intel64/libmkl_core.so
prepend-path LD_PRELOAD ${MKLROOT}/lib/intel64/libmkl_sequential.so
append-path PYTHONPATH $base/lib/python${PYTHONVER}/site-packages
Note that you may need to exchange cinderella, ${BUILDTYPE}, ${MKLROOT} and ${PYTHONVER} with your versions and possibly also the modules in prereq (check the version numbers).
Finally, it is recommended to generate a symbolic link for certain build types, e.g. serial may link to serial-vX.X.20xx-... so that module load ngsolve/serial load the correspondingly linked version of this build type, e.g.
ln -s ${BASEDIR}/modules/ngsolve/${BUILDTYPE} ${BASEDIR}/modules/ngsolve/serial
MPI parallel setup
To obtain an mpi-parallel setup only a few minor changes need to be carried out:
-
make sure you have
mpi4pyinstalled:pip install --user mpi4py -
rename
serialtoparallelormpiin the preceeding steps -
additionally load the
openmpimodule (before building):module load openmpi/gcc/64/current -
change build options, especially use
-DUSE_MUMPS=ON \ -DUSE_HYPRE=ON \ -DUSE_MPI=ON \ -DMKL_SDL=OFF \ -
Add the
openmpidependency also to your resulting module:prereq openmpi/gcc/64/current -
You may also want to install
PETSc, the simplest way to do it is viapip:pip install --user petsc petsc4py -
If you install petsc and/or petsc4py in a non-standard target (as it makes sense when setting up a module) make sure to add the corresponding path to
LD_LIBRARY_PATHin themodulefile.
Shortcut: Installation from script
In this script we provide a script that executes all the previous steps after you logged in, generated the basic directory structure and checked out the NGSolve sources[5]The steps after the log in are also contained in the script as comments.. The script also uses a slightly more sophisticated naming for the buildtype as you can see in the script, it automatically generates the module files and puts this into the modules directory and generates corresponding symlinks. Further, it is configured for the mpi-version of the installation. In case version numbers change, please make sure to update those throughout the script.
In case you have a slightly different setup (or package, ...) you may want to take the script as a starting point, so that you can easily reproduce (and share) your configuration.
Setting up an NGSolve run with the GWDG batch system (slurm)
The batch system is to be used for the actual computation. However, if you want to test your setup interactively, you may want to try an interactive session, see below..
We again suggest a simple directory structure for your simulation runs. We create the directory runs
cd $BASEDIR
mkdir -p runs
For a run we create a new directory and give it a name[6]It's not the worst idea to leave the date in the naming so that you have a simple criteria to sort your simulations.
export RUNNAME="rumpelstilzchen"
mkdir -p runs/${RUNNAME}
cd runs/${RUNNAME}
Here, we now create a file run that can be interpreted by slurm the batch system manager:
#!/bin/bash
#SBATCH -J rumpelstilzchen
#SBATCH -p medium
#SBATCH -t 12:00:00
#SBATCH -o outfile-%J
#SBATCH -N 1
#SBATCH -n 4
#SBATCH -C fas
module load ${PYTHONMODULE}
module load gcc/9.3.0
module load intel-parallel-studio/cluster.2020.4
module load ngsolve/serial
python3 ngs-example.py
Here, we set many options, like queue, max. runtime, number of nodes, number of cores, max. amount of memory needed. This relates to the options after #SBATCH. Explanations to these options are given here. The last one -C fas is important to circumvent getting IvyBridge architecture where avx support is insufficient (nodes gwdd 001-168). It forces the cluster to use computing nodes at the Faßberg.
Afterwards we load the corresponding modules that make NGSolve runable in the desired version. Note that of course ngsolve/serial should be replaced with your ${BUILDTYPE} variable. Finally, we prescribe the command(s) that should be executed in the corresponding environment, here python3 ngs-example.py.
Now, we can submit the script to the queue with
sbatch run
To check the status of (all your running) jobs call
squeue -u cinderella
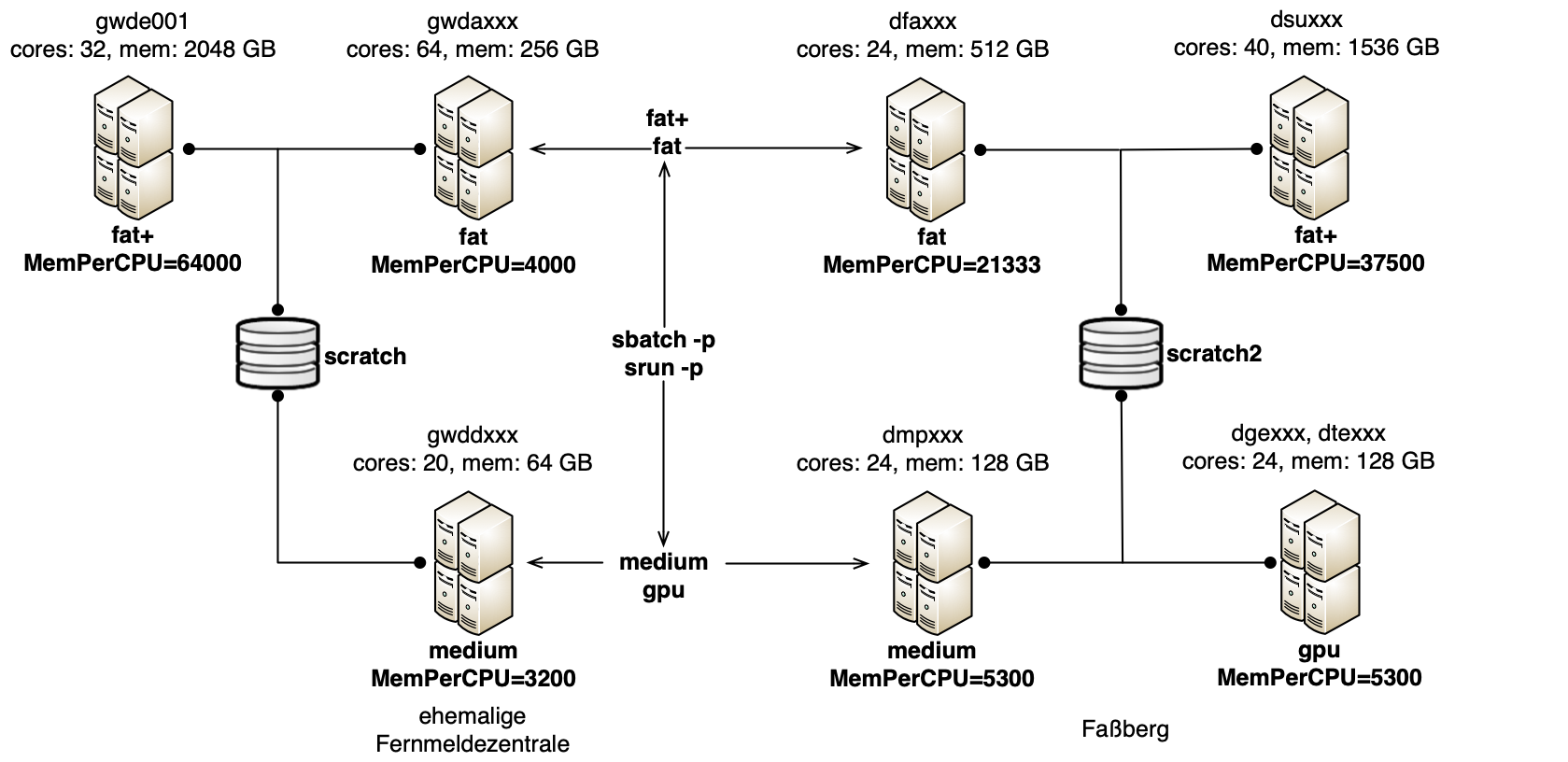
Remark: The selection of the right queue for your job is important. On the one hand the size of the job should fit the queue, on the other hand the architecture should be compatible to your built NGSolve installation. Consider also the information on the architectures involved here.
File access
To get access to the cluster filesystem you may want to use tools like sshfs, filezilla etc. Make sure that you find a convenient way to exchange files with your cluster home directory in order to process results or upload simulation configurations.
Interactive session
For debugging and testing purposes it is sometimes useful to get an interactive session. Basically, this allows to interact with NGSolve like it would be running on your Laptop.
The basic command for this is
srun -p int -c 20 -N 1 --pty bash
A detailed description is available [6]. Please note that the required modules then have to be loaded manually.
An example run (serial)
If everything went well so far and you opted for the MPI installation, you should try the following steps:
-
Download
poisson.pyfrom theNGSolvesources and put them into one directory (here we choose${BASEDIR}/runs/test)mkdir -p ${BASEDIR}/runs/test cd ${BASEDIR}/runs/test wget https://raw.githubusercontent.com/NGSolve/ngsolve/master/py_tutorials/poisson.py -
Start an interactive session:
srun -p int -c 20 -N 1 --pty bash -
In that environment load the modules as before
module load ${PYTHONMODULE} module load gcc/9.3.0 module load intel-parallel-studio/cluster.2020.4 module load ngsolve/serial -
Execute the run:
python3 poisson.py -
If the error displayed in the outpu is reasonable (e.g. < 1e-4) your test was most certainly successful.
-
You see no visualization directly from this run (as this would be in the batch system). There are at least two options to visualize results after the simulation run:
-
You can use pickling to store the result and load it later in a local installation. This can of course also be used for other processing purposes. For pickling add the following at the end of the python file:
import pickle pickle.dump (gfu, open("solution.pickle", "wb"))This will store the
GridFunction, but can also be used for other objects. Now, from a local installation (with GUI) you can draw the pickledGridFunctionwithfrom ngsolve import Draw from netgen import gui import pickle gfu = pickle.load (open("solution.pickle", "rb")) Draw(gfu) -
Alternatively, you can also use the
VTKOutput.
-
-
done. It seems that you are able to use
NGSolveon the cluster in a serial run. In the next example we explain how to test a parallelNGSolveinstallation.
An example run (MPI parallel)
If everything went well so far and you opted for the MPI installation, you should try the following steps:
-
Download
poisson_mpi.pyanddrawsolution.pyfrom theNGSolve i-tutorialon basic MPI usage and put them into one directory (here we choose${BASEDIR}/runs/mpitest)mkdir -p ${BASEDIR}/runs/mpitest cd ${BASEDIR}/runs/mpitest wget https://ngsolve.org/docu/latest/i-tutorials/unit-5a.1-mpi/poisson_mpi.py wget https://ngsolve.org/docu/latest/i-tutorials/unit-5a.1-mpi/drawsolution.py -
Start an interactive session with more nodes, here 4:
srun -p int -c 20 -N 4 --pty bash -
In that environment load the modules as before (including
openmpiandngsolve/mpi)module load ${PYTHONMODULE} module load gcc/9.3.0 module load intel-parallel-studio/cluster.2020.4 module load openmpi/gcc/64/current module load ngsolve/mpi -
Execute the MPI run:
mpirun -np 4 python3 poisson_mpi.pyThis run uses no GUI as we will not have an interactive GUI access typically when running in the batch system, especially not with MPI. To inspect the simulation results however, the result is pickled and due to
netgen.meshing.SetParallelPickling(True)the results are moved to the master process which allows to have a serial view on the result later on. If the result is still to large to be processed (or you like that option for other reasons), you can also use theVTKOutput which is carried out in parallel and usingParaViewcan also be used for visualization in a parallel mode. -
Next, get access to the
drawsolution.pyandsolution.pickle0from a localNGSolveinstallation with GUI (either via mounting the cluster filesystem or by copying the file) and execute the formernetgen drawsolution.pyYou should now get a visualization of the result that was obtained in parallel.
-
done.
References
[3] GWDG HPC cluster introduction
[4] Slurm scripts on GWDG HPC cluster
